Recently, the research team from Department of Ocean Science and Otto Poon Center for Climate Resilience and Sustainability at Hong Kong University of Science and Technology, and the Marine Molecular Ecology and Chemical Ecology Team of the Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) have published a research paper titled "DOO: Integrated Multi-Omics Resources for Deep Ocean Organisms" in Nucleic Acids Research, a top journal in the field of international bioinformatics databases.The paper was co-correspondenced by Professor Pei-yuan Qian and Professor Longjun Wu, with Dr. Jiajie She, a postdoctoral researcher, serving as the first author. Guangzhou Marine Laboratory is the first-affiliated institution.As the world’s first and largest multi-omics database for deep-sea organisms, theDOO (Deep Ocean Omics) database (https://DeepOceanOmics.org) is the world’s first and largest multi-omics database dedicated to deep-sea organisms. It aims to build an one-stop platform that integrates multi-omics data, provides customized analysis toolkits, and supports cross-species comparative and evolutionary research. It promotes the research and application of deep-sea biology, adaptive mechanisms to extreme environments, and deep-sea biological resources.
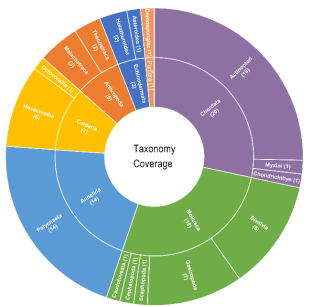
Figure 1. Overview of Taxonomy coverage of deep-sea organisms across phyla and classes in DOO
The deep ocean, as the largest ecosystem on the planet, represents one of the final unexplored frontiers on Earth, harbouring unique ecosystems and extraordinary biodiversity. The synergistic interplay of abyssal stressors, including high pressure, oxygen deficiency, perpetual darkness, low temperature and nutrient limitation, imposes profound physiological constraints on deep ocean organisms. In recent years, breakthroughs in high-throughput sequencing technologies have led to major progress in multi-omics research on deep-sea organisms, revealing unique adaptations at genetic, metabolic, and symbiotic mechanisms. However, the lack of integrated resources, standardized data, and dedicated analytical tools has hindered effective consolidation and exploration of these multi-omics datasets.
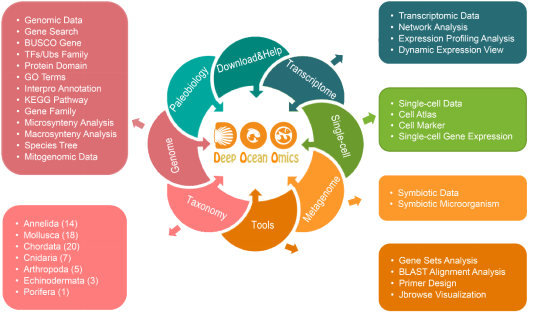
Figure 2. Database architecture and multi-omics data visualization and functional analysis modules
To address this gap, the research team manually collected and integrated multi-omics data from 68 deep-sea animal species, including 72 genomes, 950 transcriptomes, 1,112 metagenomes, and 15 single-cell transcriptomes. The database covers species across 7 phyla (Mollusca, Annelida, Arthropoda, Chordata, Cnidaria, Echinodermata, and Porifera) from diverse deep-sea habitats such as cold seeps, hydrothermal vents, and seamounts. It also incorporates 1,413 fossil records to support evolutionary analyses of environmental adaptation strategies in deep-sea organisms. This makes DOO the most comprehensive multi-omics platform for deep-sea species in terms of both taxonomic breadth and data dimensions. Additionally, DOO integrates multiple functional modules, including Gene and Genome Module (gene structure and functional annotation, transcription factors, ubiquitin families, transposons, gene families, etc.), Functional Genomics Analysis Module (gene co-expression networks, dynamic network view, single-cell data visualization, and metagenomic analysis), and Evolutionary and Comparative Genomics Module (pan-geneset analysis, micro- and macro-synteny analysis among deep-sea species, ancestral karyotype reconstruction, and phylogenetic tree construction). The platform also incorporates multi-dimensional data into a customized Jbrowse genome browser, enabling integrated visualization and analysis. Since its launch, DOO has attracted over 900 visits from users in 28 countries.
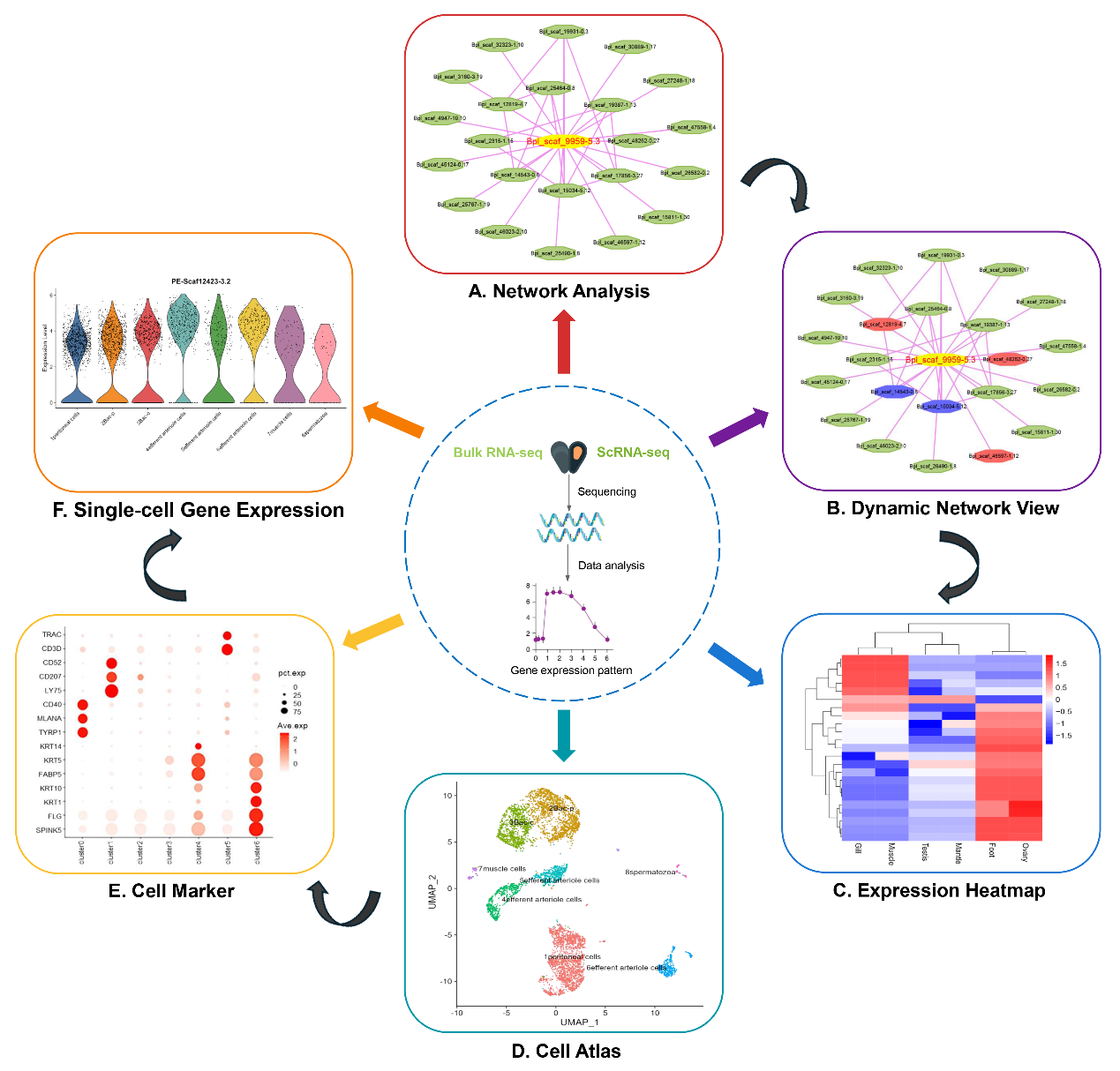
Figure 3. Visualization of bulk and single-cell transcriptomes in DOO
The work was funded by grants (2021HJ01, HJRC2022001, and SMSEGL24SC01) of the Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), and Otto Poon Center for Climate Resilience and Sustainability (CCRS25SC01), the Research Grant Council (RGC) of the Hong Kong Special Administrative Region, Outstanding Young Scholars Scheme of the Research Grants Council of Hong Kong (26104824) and the Science and Technology Innovation Committee of Shenzhen (JCYJ20220530151207016).
Attachment download:
